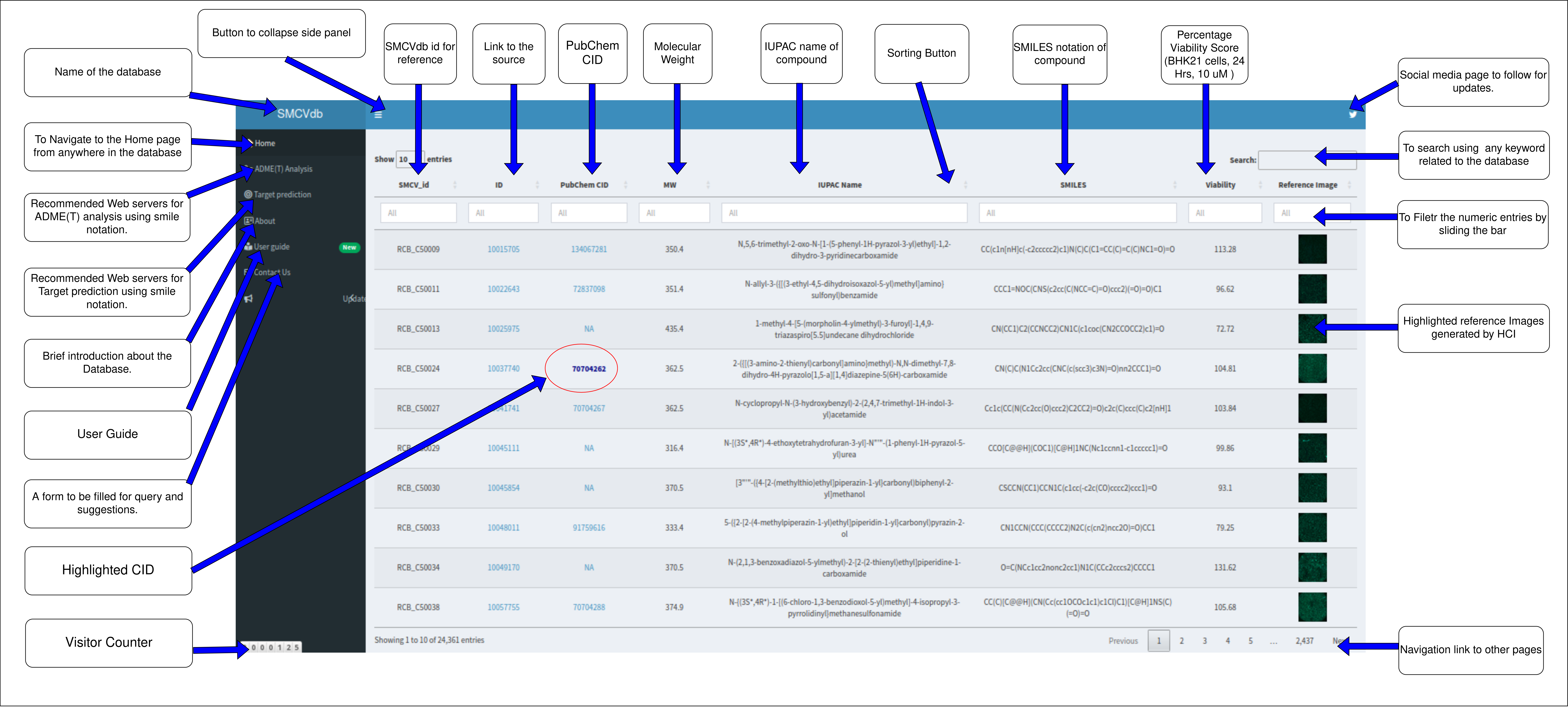
Recommended Web servers for ADME(T) analysis
References
1. Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., Yin, M., Zeng, X., Wu, C., Lu, A., Chen, X., Hou, T., & Cao, D. (2021). ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic acids research, 49(W1), W5–W14. https://doi.org/10.1093/nar/gkab255
2. Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports, 7, 42717. https://doi.org/10.1038/srep42717
3. Yang, H., Lou, C., Sun, L., Li, J., Cai, Y., Wang, Z., Li, W., Liu, G., & Tang, Y. (2019). admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics (Oxford, England), 35(6), 1067–1069. https://doi.org/10.1093/bioinformatics/bty707
Recommended Web servers for Target prediction
References
1. Gfeller, D., Grosdidier, A., Wirth, M., Daina, A., Michielin, O., & Zoete, V. (2014). SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic acids research, 42(Web Server issue), W32–W38. https://doi.org/10.1093/nar/gku293
About SMCVdb
Welcome to SMCVdb, your comprehensive resource for accessing valuable information on small molecule compounds and their effects on cell viability. The acronym stands for "Small Molecule Cell Viability Database," reflecting our commitment to aiding researchers, scientists, clinicians, and a wider audience in their endeavors related to drug discovery and development.
SMCVDB is meticulously designed to serve as a repository of essential cell viability data derived from high content imaging techniques. Our primary goal is to facilitate the exploration of small molecules' impact on cell viability. Moreover, we go beyond by providing SMILES notations for the compounds, enabling users to seamlessly locate targets and perform ADMEt (Absorption, Distribution, Metabolism, Excretion, and Toxicity) analyses through recommended servers. By streamlining these processes, we aim to accelerate drug design and development, ultimately contributing to the advancement of medical research.
Cite
Pandey, A. D., Sharma, G., Sharma, A., Vrati, S., & Nair, D. T. (2024). SMCVdb: a database of experimental cellular toxicity information for drug candidate molecules. Database : the journal of biological databases and curation, 2024, baae100. https://doi.org/10.1093/database/baae100.